

Splicing of precursor messenger RNA (pre-mRNA) is a ubiquitous hallmark of gene expression in all eukaryotes. Within the cell, splicing occurs simultaneously with pre-mRNA synthesis by RNA polymerase II (Pol II) and is mechanistically coupled to transcription. The choice of the splice sites is not always unique, as a single gene can give rise to multiple functionally distinct mRNA isoforms through alternative splicing. In fact, more than 95% of human genes are alternatively spliced, massively expanding the coding potential of our genome. Dysregulation of alternative splicing contributes to cancer pathogenesis and is under study as a biomarker of diseases. Our research aims to understand the mechanism governing alternative splice site selection and the crosstalk between the transcription and splicing machineries.

We recently determined the cryo-EM structure of the transcribing Pol II in complex with U1 snRNP, the first spliceosomal building block that engages pre-mRNA. The structure revealed for the first time a direct protein interaction between the transcription and splicing machineries within a large supercomplex and provided insights into how functional pairing of distant intron ends and spliceosome assembly can occur on the Pol II surface.
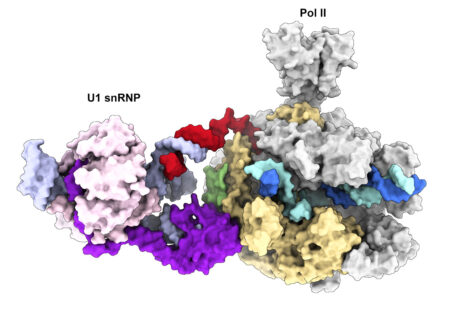
Our lab employs a multidisciplinary approach combining biochemical reconstitution, functional assays, structural biology and in vivo RNA sequencing to define the molecular basis of transcription-coupled alternative splicing. Our long-term research goal is to understand how the transcription and splicing machineries functionally interact to regulate gene expression.
Selected Papers
- Zhang, L., Gordiyenko, Y., Morgan, T., Franco, C., Tufegdžić Vidaković, A., Zhang, S. (2025)
Structural basis of RECQL5-induced RNA polymerase II transcription braking and subsequent reactivation.
Nat Struct Mol Biol - Zhang, L., Batters, C., Aibara, S., Gordiyenko, Y., Žumer, K., Schmitzová, J., Maier, K., Cramer, P., Zhang, S. (2025)
Structure of a transcribing Pol II-DSIF-SPT6-U1 snRNP complex.
Nat Commun 16(1): 5823 - Zhang S, Aibara S, Vos S, Agafonov D, Lührmann R, Cramer P. (2021)
Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science 371: 305-309. - Zhang S, Tischer T & Barford D. (2019)
Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nature Communications 10: 3863. - Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, Skehel M & Barford D. (2016)
Molecular mechanism of APC/C activation by mitotic phosphorylation.
Nature 533: 260-264. - Erzberger JP, Stengel F, Pellarin R, Zhang S, Schäfer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, Ban N. (2014)
Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex.
Cell 158(5): 1123-35.
Group Members
- Jack Bowden
- Yuliya Gordiyenko
- Pei Wang
- Helen Zhang